Some diagnostics for a fitted qgam model
check.qgam.RdTakes a fitted gam object produced by qgam() and produces some diagnostic information
about the fitting procedure and results. It is partially based on mgcv::gam.check.
# S3 method for qgam check(obj, nbin = 10, lev = 0.05, ...)
Arguments
| obj | the output of a |
|---|---|
| nbin | number of bins used in the internal call to |
| lev | the significance levels used by |
| ... | extra arguments to be passed to |
Value
Simply produces some plots and prints out some diagnostics.
Details
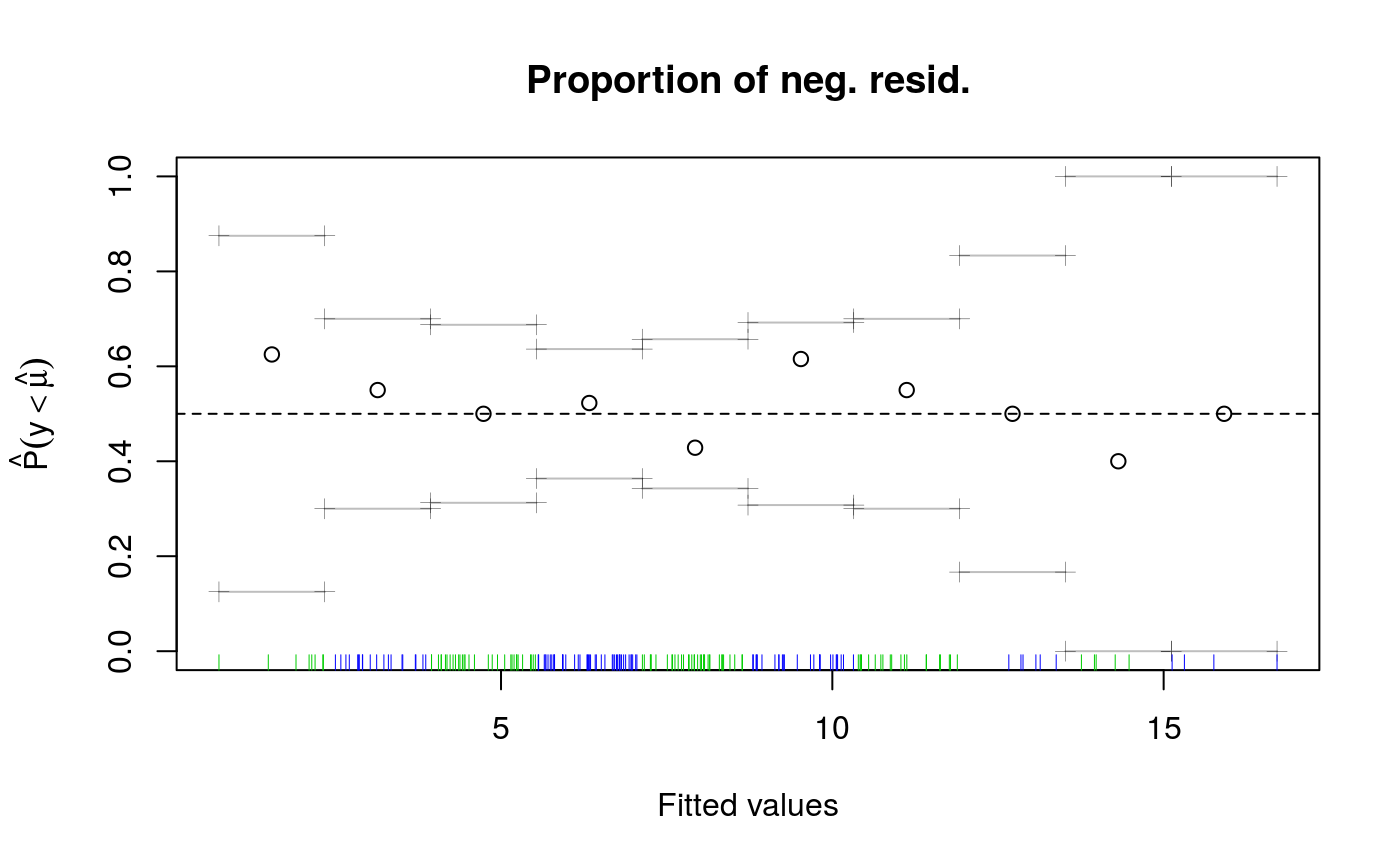
This function provides two plots. The first shows how the number of responses falling below the fitted
quantile (y-axis) changes with the fitted quantile (x-axis). To be clear: if the quantile is fixed to, say, 0.5
we expect 50% of the responses to fall below the fit. See ?cqcheck() for details. The second plot related
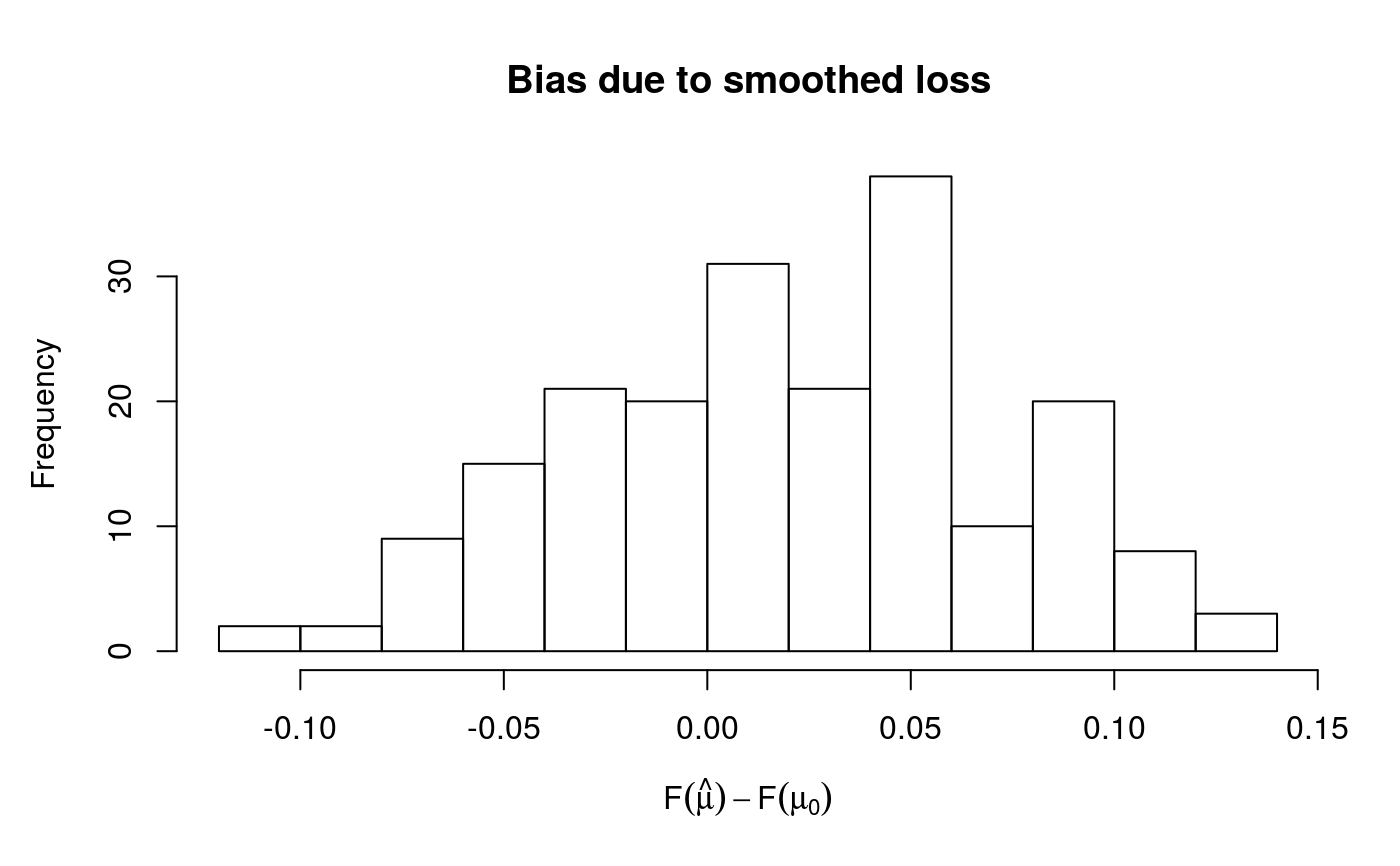
to |F(hat(mu)) - F(mu0)|, which is the absolute bias attributable to the fact that qgam is using
a smoothed version of the pinball-loss. The absolute bias is evaluated at each observation, and an histogram
is produced. See Fasiolo et al. (2017) for details. The function also prints out the integrated absolute bias,
and the proportion of observations lying below the regression line. It also provides some convergence
diagnostics (regarding the optimization), which are the same as in mgcv::gam.check.
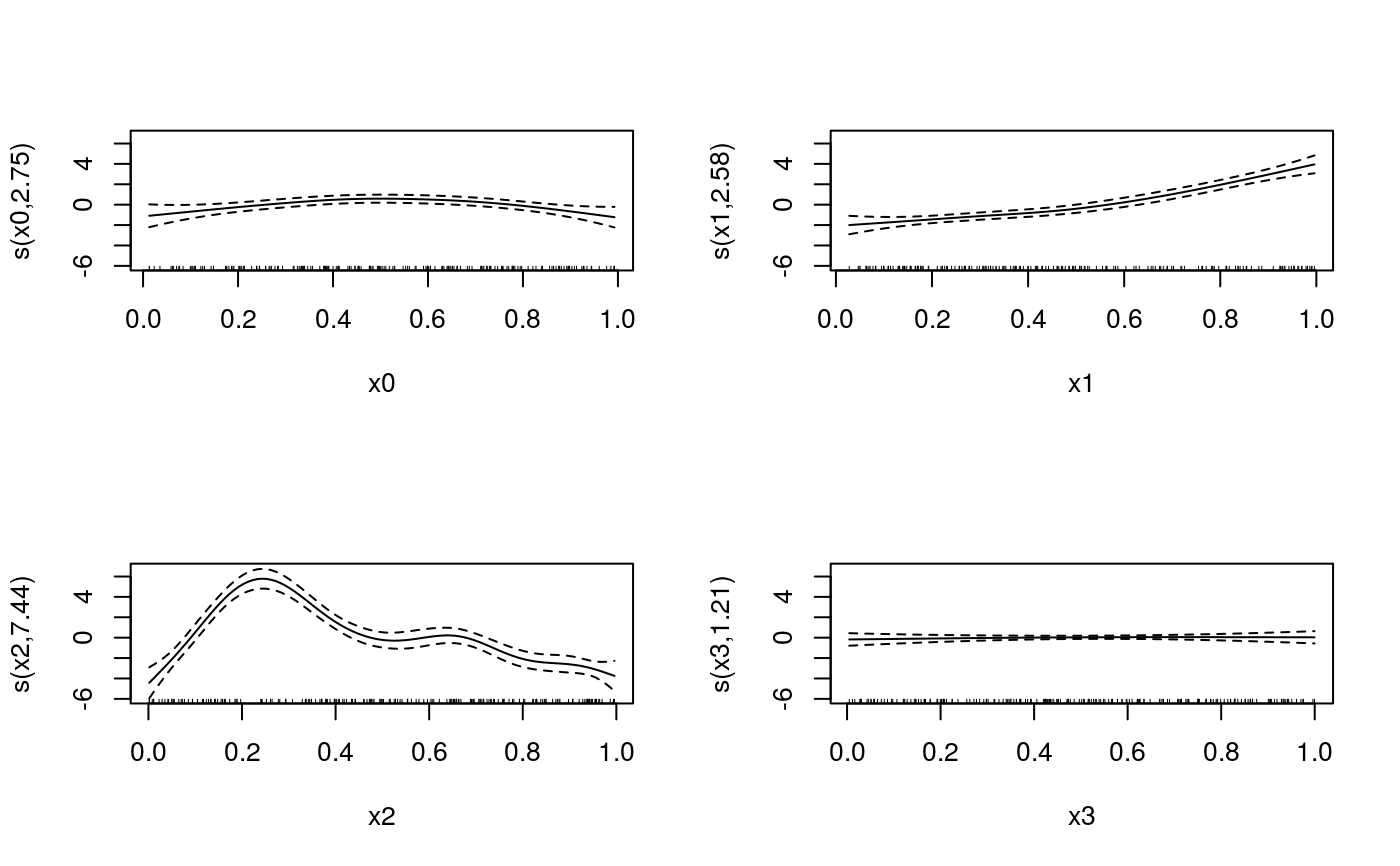
It reports also the maximum (k') and the selected degrees of freedom of each smooth term.
References
Fasiolo, M., Goude, Y., Nedellec, R. and Wood, S. N. (2017). Fast calibrated additive quantile regression. Available at https://arxiv.org/abs/1707.03307.
Examples
#> Gu & Wahba 4 term additive model#> Estimating learning rate. Each dot corresponds to a loss evaluation. #> qu = 0.5...............donecheck.qgam(b, pch=19, cex=.3)#> Theor. proportion of neg. resid.: 0.5 Actual proportion: 0.52 #> Integrated absolute bias |F(mu) - F(mu0)| = 0.04500293 #> Method: REML Optimizer: outer newton #> full convergence after 6 iterations. #> Gradient range [-0.0001503744,2.710312e-05] #> (score 454.5555 & scale 1). #> Hessian positive definite, eigenvalue range [0.02110521,2.627179]. #> Model rank = 37 / 37 #> #> Basis dimension (k) check: if edf is close too k' (maximum possible edf) #> it might be worth increasing k. #> #> k' edf #> s(x0) 9 2.75 #> s(x1) 9 2.58 #> s(x2) 9 7.44 #> s(x3) 9 1.21