Tuning the learning rate for Gibbs posterior
tuneLearn.RdThe learning rate (sigma) of the Gibbs posterior is tuned either by calibrating the credible intervals for the fitted curve, or by minimizing the pinball loss on out-of-sample data. This is done by bootrapping or by k-fold cross-validation. Here the calibration loss function is evaluated on a grid of values provided by the user.
tuneLearn(form, data, lsig, qu, err = NULL, multicore = !is.null(cluster), cluster = NULL, ncores = detectCores() - 1, paropts = list(), control = list(), argGam = NULL)
Arguments
| form | A GAM formula, or a list of formulae. See ?mgcv::gam details. |
|---|---|
| data | A data frame or list containing the model response variable and covariates required by the formula. By default the variables are taken from environment(formula): typically the environment from which gam is called. |
| lsig | A vector of value of the log learning rate (log(sigma)) over which the calibration loss function is evaluated. |
| qu | The quantile of interest. Should be in (0, 1). |
| err | An upper bound on the error of the estimated quantile curve. Should be in (0, 1).
Since qgam v1.3 it is selected automatically, using the methods of Fasiolo et al. (2017).
The old default was |
| multicore | If TRUE the calibration will happen in parallel. |
| cluster | An object of class |
| ncores | Number of cores used. Relevant if |
| paropts | a list of additional options passed into the foreach function when parallel computation is enabled. This is important if (for example) your code relies on external data or packages: use the .export and .packages arguments to supply them so that all cluster nodes have the correct environment set up for computing. |
| control | A list of control parameters for
|
| argGam | A list of parameters to be passed to |
Value
A list with entries:
lsig= the value of log(sigma) resulting in the lowest loss.loss= vector containing the value of the calibration loss function corresponding to each value of log(sigma).edf= a matrix where the first colums contain the log(sigma) sequence, and the remaining columns contain the corresponding effective degrees of freedom of each smooth.convProb= a logical vector indicating, for each value of log(sigma), whether the outer optimization which estimates the smoothing parameters has encountered convergence issues.FALSEmeans no problem.
References
Fasiolo, M., Goude, Y., Nedellec, R. and Wood, S. N. (2017). Fast calibrated additive quantile regression. Available at https://arxiv.org/abs/1707.03307.
Examples
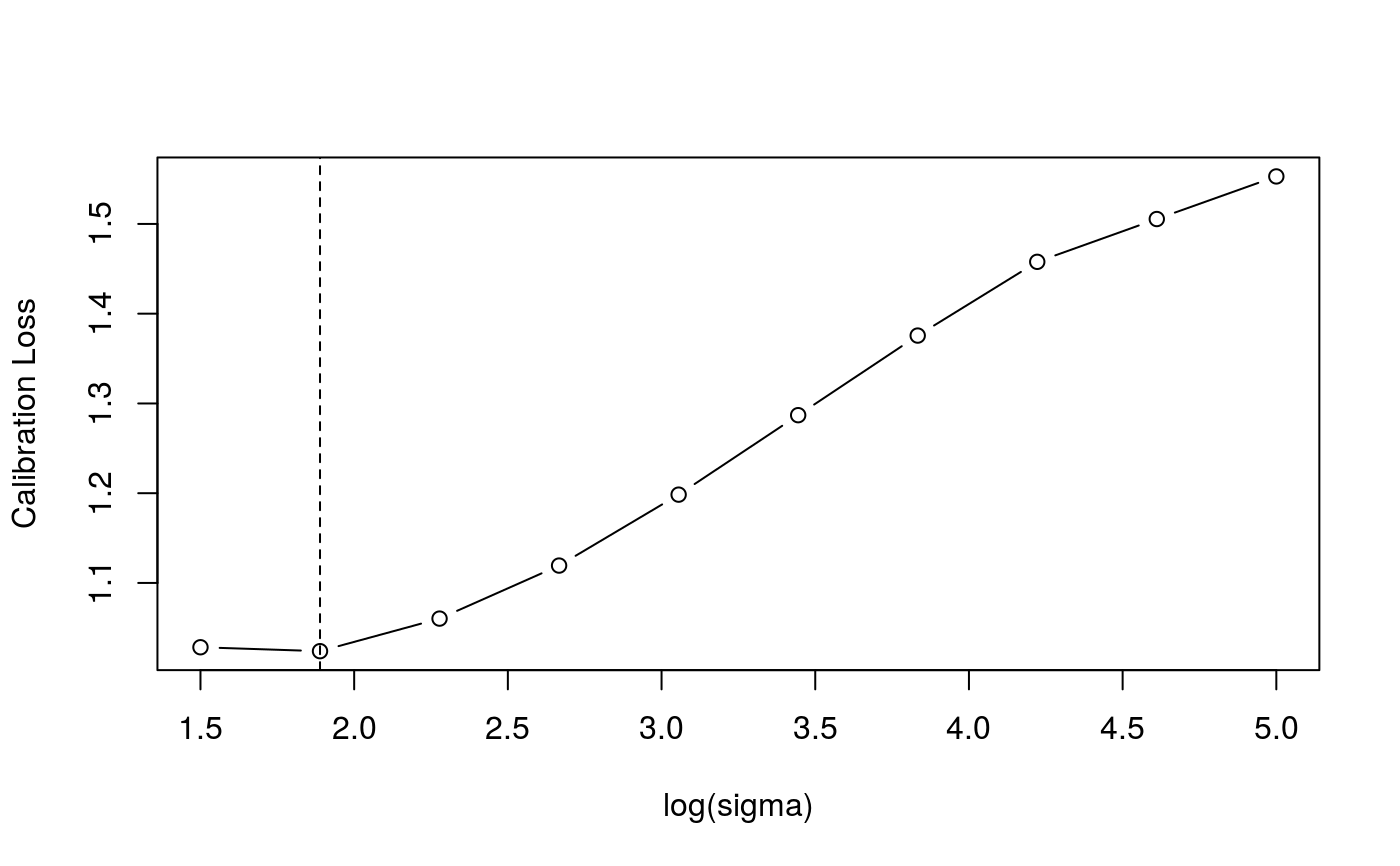
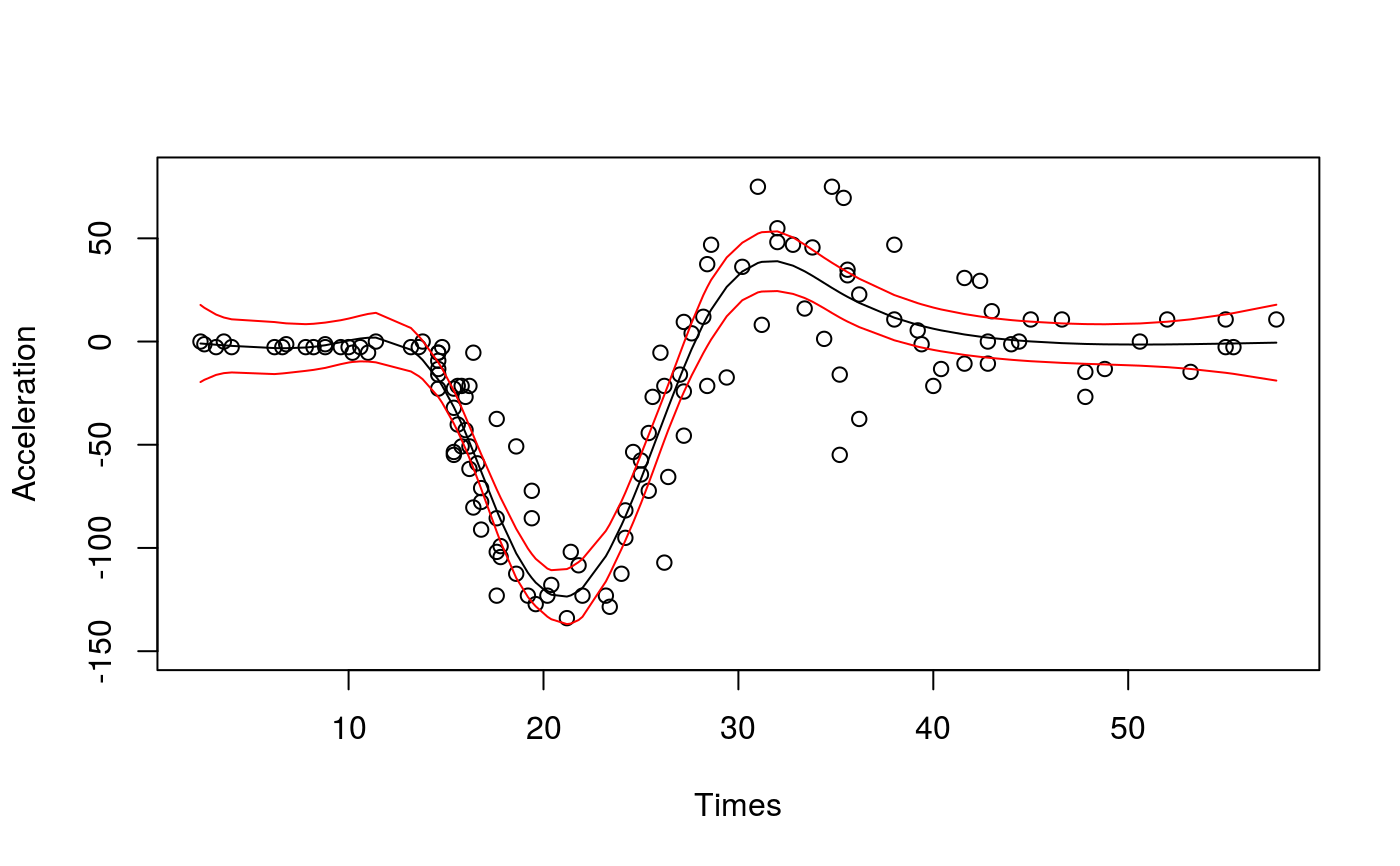
library(qgam); library(MASS) # Calibrate learning rate on a grid set.seed(41444) sigSeq <- seq(1.5, 5, length.out = 10) closs <- tuneLearn(form = accel~s(times,k=20,bs="ad"), data = mcycle, lsig = sigSeq, qu = 0.5) plot(sigSeq, closs$loss, type = "b", ylab = "Calibration Loss", xlab = "log(sigma)")# Fit using the best sigma fit <- qgam(accel~s(times,k=20,bs="ad"), data = mcycle, qu = 0.5, lsig = best) summary(fit)#> #> Family: elf #> Link function: identity #> #> Formula: #> accel ~ s(times, k = 20, bs = "ad") #> #> Parametric coefficients: #> Estimate Std. Error z value Pr(>|z|) #> (Intercept) -25.20 1.88 -13.4 <2e-16 *** #> --- #> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 #> #> Approximate significance of smooth terms: #> edf Ref.df Chi.sq p-value #> s(times) 8.73 10.11 531.3 <2e-16 *** #> --- #> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 #> #> R-sq.(adj) = 0.782 Deviance explained = 71.5% #> -REML = 608.79 Scale est. = 1 n = 133pred <- predict(fit, se=TRUE) plot(mcycle$times, mcycle$accel, xlab = "Times", ylab = "Acceleration", ylim = c(-150, 80))