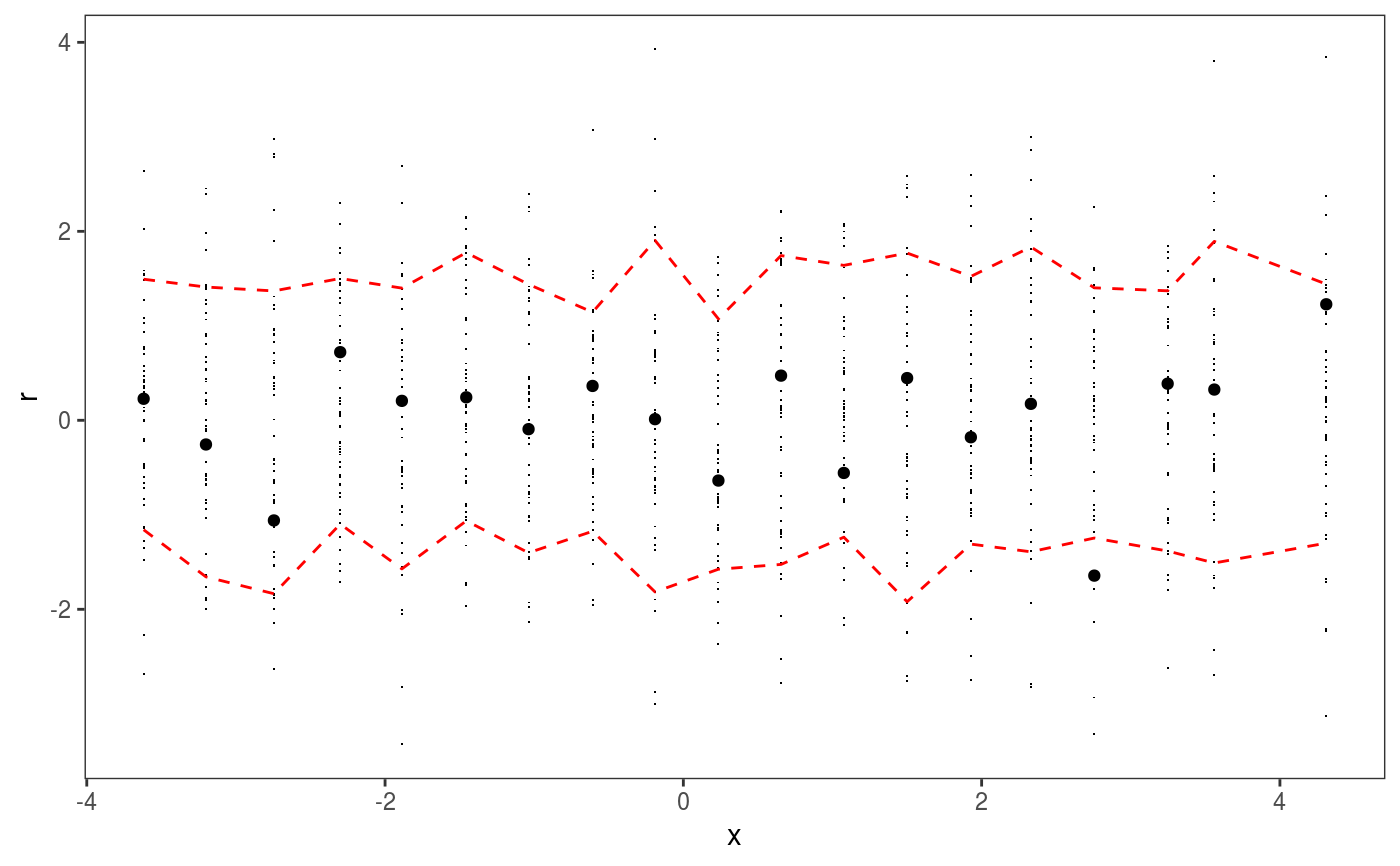
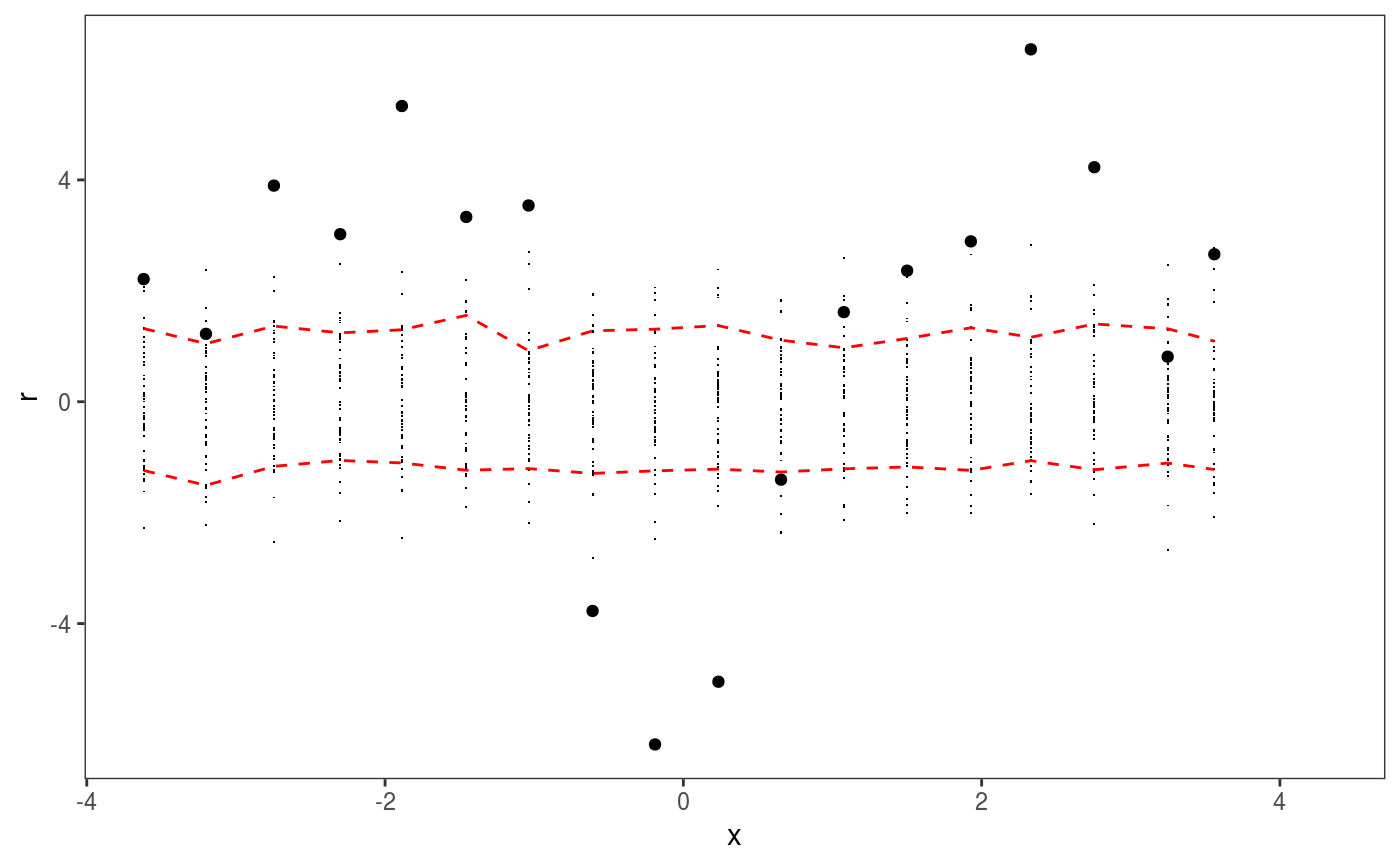
This layer bins the residuals, r, according to the value of the corresponding covariate, x. Then the residuals in each bin are summarized using a scalar-valued statistic. Confidence intervals for the statistic corresponding to each bin can be obtained by simulating residuals from the fitted GAM model, binning and summarizing them. Mainly useful in conjuction with check1D.
l_gridCheck1D( gridFun = NULL, n = 20, level = 0.8, stand = "none", showReps = TRUE, showObs = TRUE, ... )
Arguments
| gridFun | scalar-valued function used to summarize the residuals in each bin.
It takes a vector as input. By default it is
|
|---|---|
| n | number of grid intervals along the relevant covariate. |
| level | the level of the confidence intervals (e.g. 0.9 means 90% intervals). |
| stand | if "none" the residuals in each bin are transformed by |
| showReps | if |
| showObs | if |
| ... | graphical arguments to be passed to |
Value
An object of class gamLayer
Examples
library(mgcViz); set.seed(4124) n <- 1e4 x <- rnorm(n); y <- rnorm(n); # Residuals are heteroscedastic w.r.t. x ob <- (x)^2 + (y)^2 + (0.2*abs(x) + 1) * rnorm(n) b <- bam(ob ~ s(x,k=30) + s(y, k=30), discrete = TRUE) b <- getViz(b, nsim = 50) # Don't see much by looking at mean check1D(b, "x") + l_gridCheck1D()# Heteroscedasticity clearly visible here check1D(b, "x") + l_gridCheck1D(gridFun = sd, stand = "sc") # <- we are scaling and centering# Last point on the right of the rug seems to indicate that a bin is missing. # It is not an error, only on observation falls in that bin, hence the # standard deviation is not defined there.