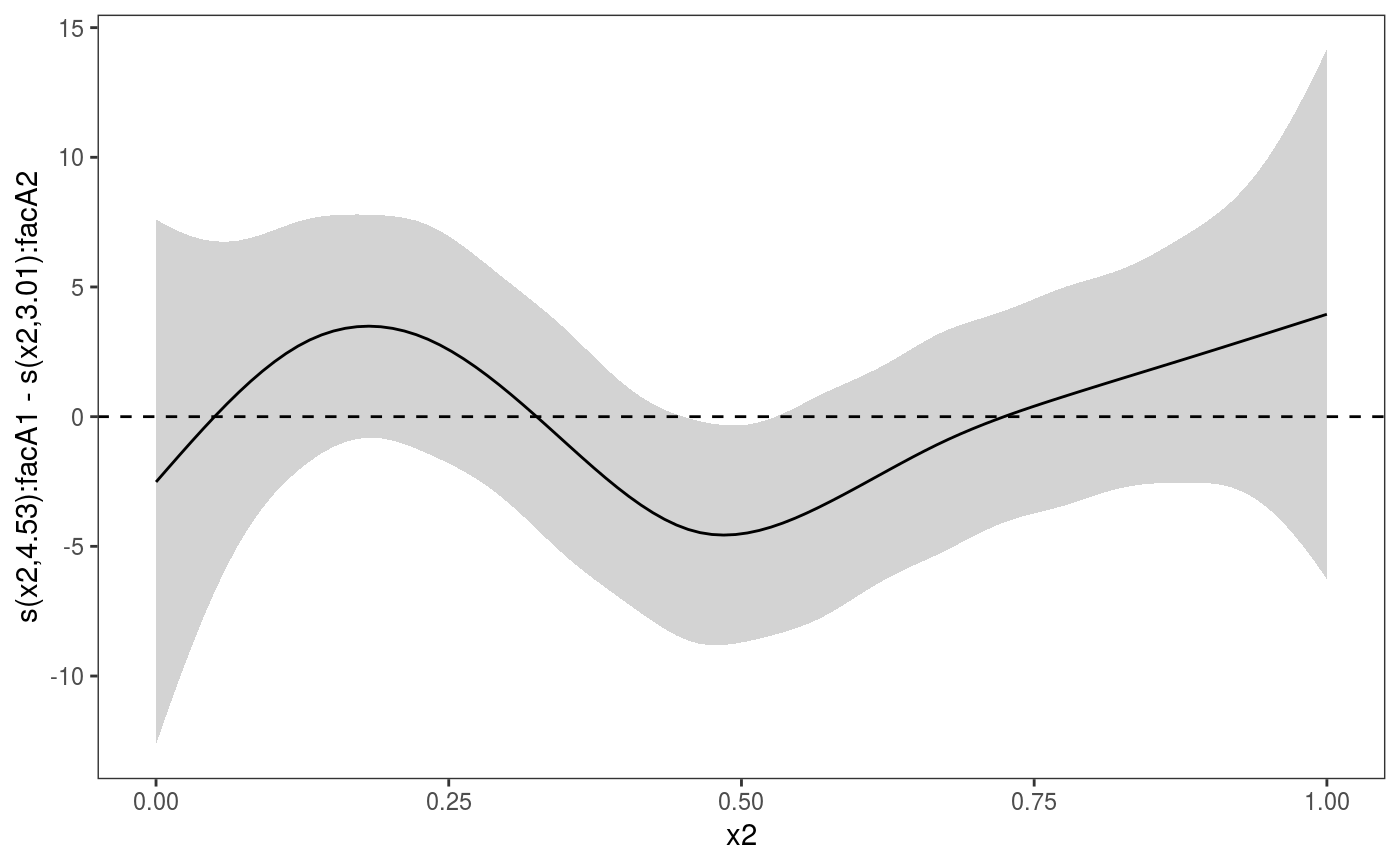
Plotting differences between two 1D smooth effects
Source:R/plotDiff_mgcv_smooth_1D.R
plotDiff.mgcv.smooth.1D.RdThis method can be used to plot the difference between two 1D smooth effects. Mainly meant to be used with by-factor smooths.
# S3 method for mgcv.smooth.1D plotDiff(s1, s2, n = 100, trans = identity, unconditional = FALSE, ...)
Arguments
| s1 | a smooth effect object, extracted using mgcViz::sm. |
|---|---|
| s2 | another smooth effect object. |
| n | number of grid points used to compute main effects and c.i. lines. For a nice smooth plot this needs to be several times the estimated degrees of freedom for the smooth. |
| trans | monotonic function to apply to the smooth and residuals, before plotting. Monotonicity is not checked. |
| unconditional | if |
| ... | currently unused. |
Value
An objects of class plotSmooth.
Details
Let sd be the difference between the fitted smooths, that is: sd = s1 - s2. sd is a vector of length n, and its covariance matrix is Cov(sd) = X1\ where: X1 (X2) and Sig11 (Sig22) are the design matrix and the covariance matrix of the coefficients of s1 (s2), while Sig12 is the cross-covariance matrix between the coefficients of s1 and s2. To get the confidence intervals we need only diag(Cov(sd)), which here is calculated efficiently (without computing the whole of Cov(sd)).
References
Marra, G and S.N. Wood (2012) Coverage Properties of Confidence Intervals for Generalized Additive Model Components. Scandinavian Journal of Statistics.
Examples
# Simulate data and add factors uncorrelated to the response library(mgcViz) set.seed(6898) dat <- gamSim(1,n=1500,dist="normal",scale=20)#> Gu & Wahba 4 term additive modeldat$fac <- as.factor( sample(c("A1", "A2", "A3"), nrow(dat), replace = TRUE) ) dat$logi <- as.logical( sample(c(TRUE, FALSE), nrow(dat), replace = TRUE) ) bs <- "cr"; k <- 12 b <- gam(y ~ s(x2,bs=bs,by = fac), data=dat) o <- getViz(b, nsim = 0) # Extract the smooths correspoding to "A1" and "A2" and plot their differences # with credible intervals plotDiff(s1 = sm(o, 1), s2 = sm(o, 2)) + l_ciPoly() + l_fitLine() + geom_hline(yintercept = 0, linetype = 2)