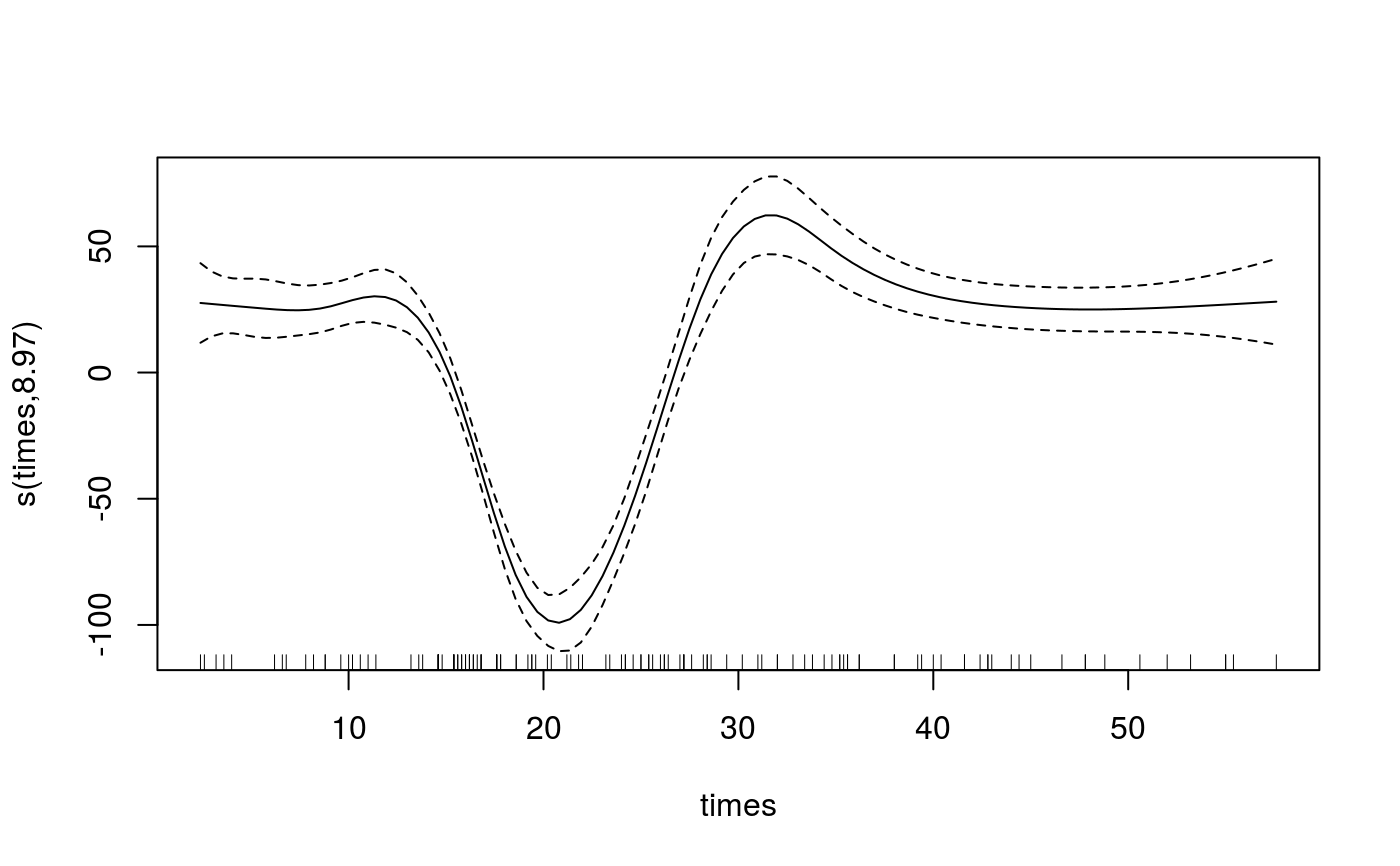
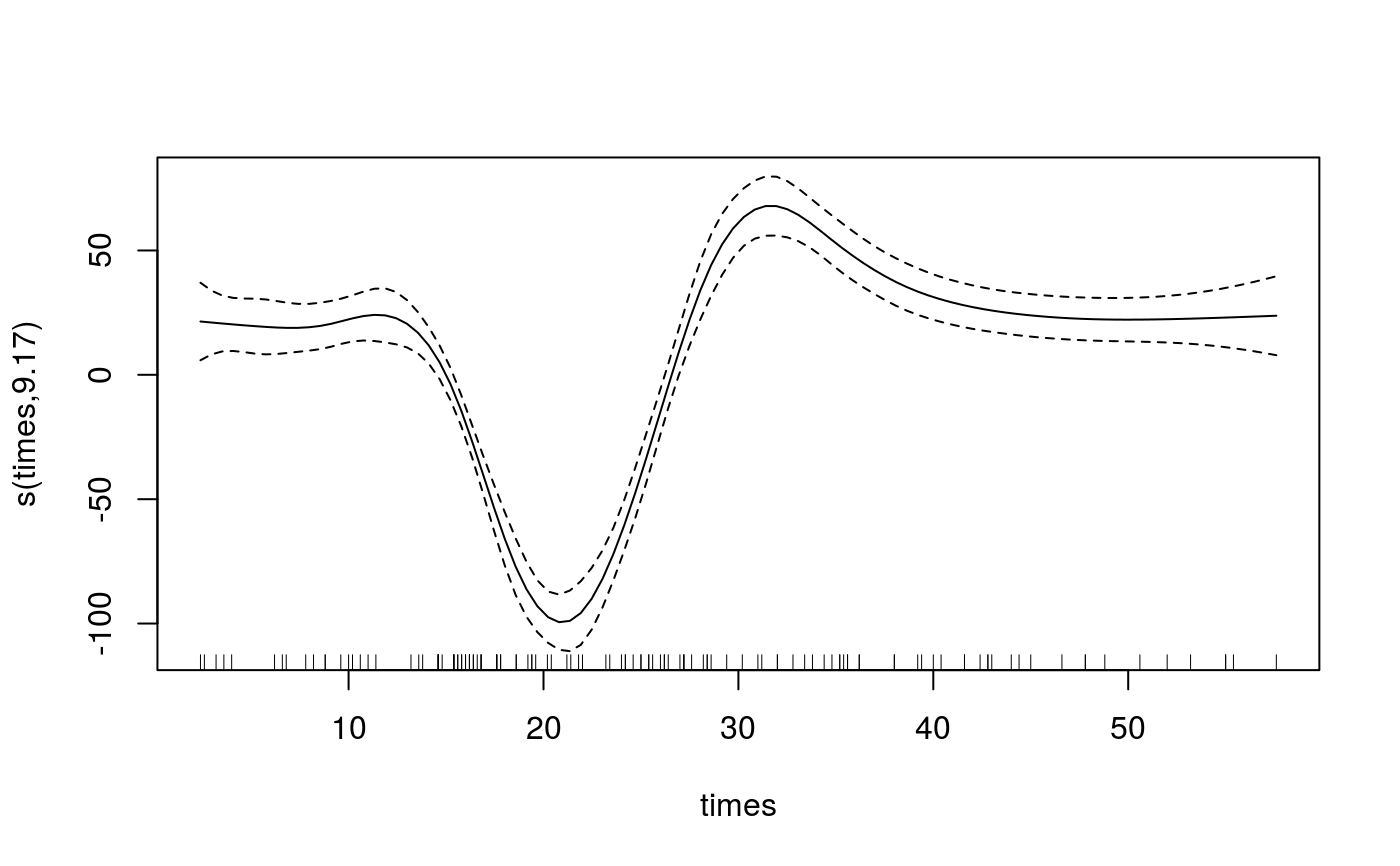
Manipulating the output of mqgam
qdo.RdContrary to qgam, mqgam does not output a standard gamObject, hence
methods such as predict.gam or plot.gam cannot be used directly. qdo
provides a simple wrapper for such methods.
qdo(obj, qu = NULL, fun = I, ...)
Arguments
| obj | the output of a |
|---|---|
| qu | A vector whose elements must be in (0, 1). Each element indicates a quantile of interest,
which should be an element of |
| fun | The method or function that we want to use on the |
| ... | Additional arguments to be passed to |
Value
A list where the i-th entry is the output of fun (whatever that is) corresponding to quantile qu[i].
Examples
library(qgam); library(MASS) quSeq <- c(0.4, 0.6) set.seed(737) fit <- mqgam(accel~s(times, k=20, bs="ad"), data = mcycle, qu = quSeq)#> Estimating learning rate. Each dot corresponds to a loss evaluation. #> qu = 0.4..........done #> qu = 0.6...........doneqdo(fit, 0.4, summary)#> #> Family: elf #> Link function: identity #> #> Formula: #> accel ~ s(times, k = 20, bs = "ad") #> #> Parametric coefficients: #> Estimate Std. Error z value Pr(>|z|) #> (Intercept) -31.181 1.832 -17.02 <2e-16 *** #> --- #> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 #> #> Approximate significance of smooth terms: #> edf Ref.df Chi.sq p-value #> s(times) 8.968 10.35 666.4 <2e-16 *** #> --- #> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 #> #> R-sq.(adj) = 0.781 Deviance explained = 69.9% #> -REML = 609.58 Scale est. = 1 n = 133