Fit multiple smooth additive quantile regression models
mqgam.RdThis function fits a smooth additive regression model to several quantiles.
mqgam(form, data, qu, lsig = NULL, err = NULL, multicore = !is.null(cluster), cluster = NULL, ncores = detectCores() - 1, paropts = list(), control = list(), argGam = NULL)
Arguments
| form | A GAM formula, or a list of formulae. See ?mgcv::gam details. |
|---|---|
| data | A data frame or list containing the model response variable and covariates required by the formula. By default the variables are taken from environment(formula): typically the environment from which gam is called. |
| qu | A vectors of quantiles of interest. Each entry should be in (0, 1). |
| lsig | The value of the log learning rate used to create the Gibbs posterior. By defauls |
| err | An upper bound on the error of the estimated quantile curve. Should be in (0, 1). If it is a vector, it should be of the
same length of |
| multicore | If TRUE the calibration will happen in parallel. |
| cluster | An object of class |
| ncores | Number of cores used. Relevant if |
| paropts | a list of additional options passed into the foreach function when parallel computation is enabled. This is important if (for example) your code relies on external data or packages: use the .export and .packages arguments to supply them so that all cluster nodes have the correct environment set up for computing. |
| control | A list of control parameters. The only one relevant here is |
| argGam | A list of parameters to be passed to |
Value
A list with entries:
fit= agamObject, one for each entry ofqu. Notice that the slotsmodelandsmoothof each object has been removed to save memory. See?gamObject.model= themodelslot of thegamObjects in thefitslot. This is the same for every fit, hence only one copy is stored.smooth= thesmoothslot of thegamObjects in thefitslot. This is the same for every fit, hence only one copy is stored.calibr= a list which is the output of an internal call totuneLearnFast, which is used for calibrating the learning rate. See?tuneLearnFastfor details.
References
Fasiolo, M., Goude, Y., Nedellec, R. and Wood, S. N. (2017). Fast calibrated additive quantile regression. Available at https://arxiv.org/abs/1707.03307.
Examples
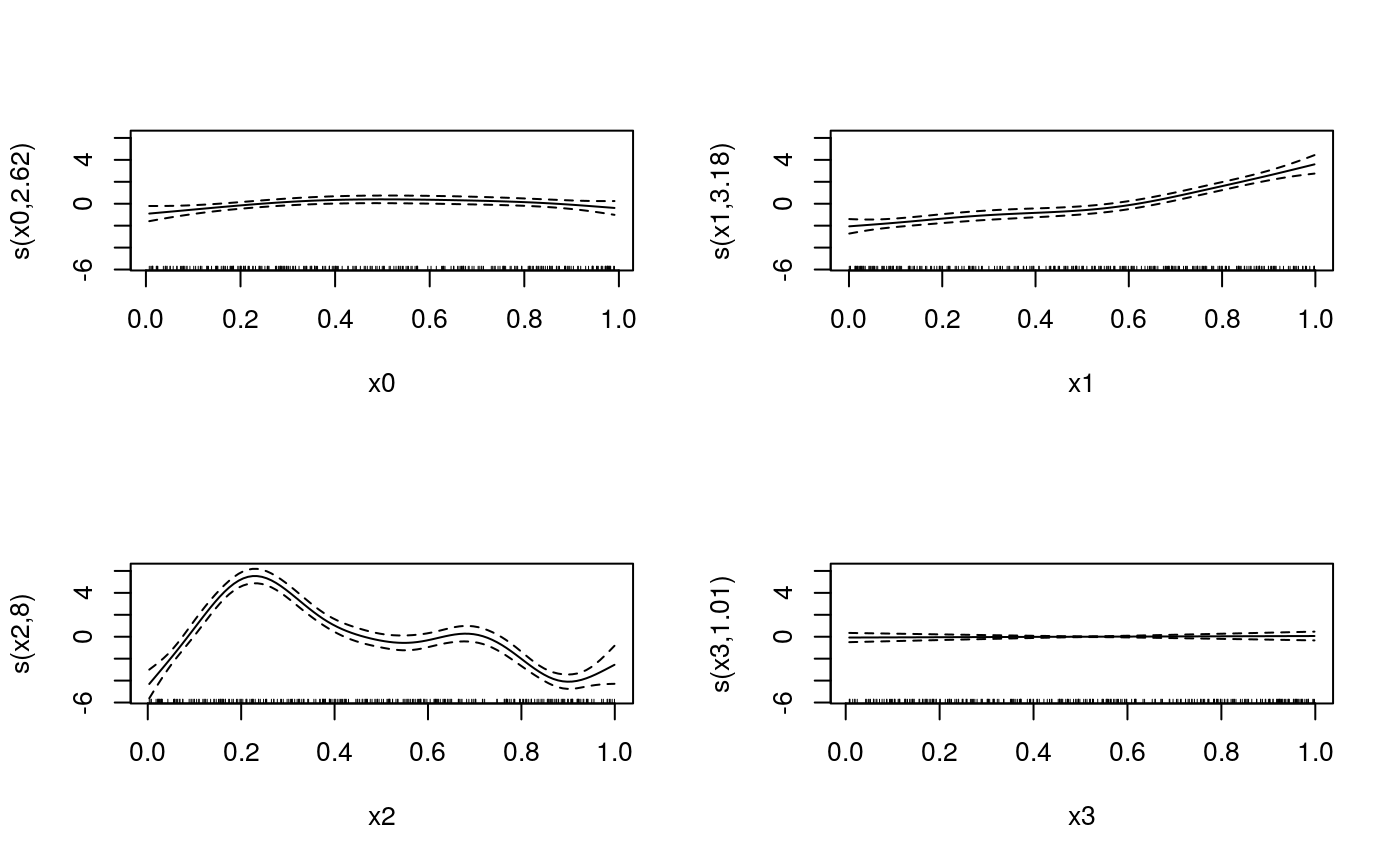
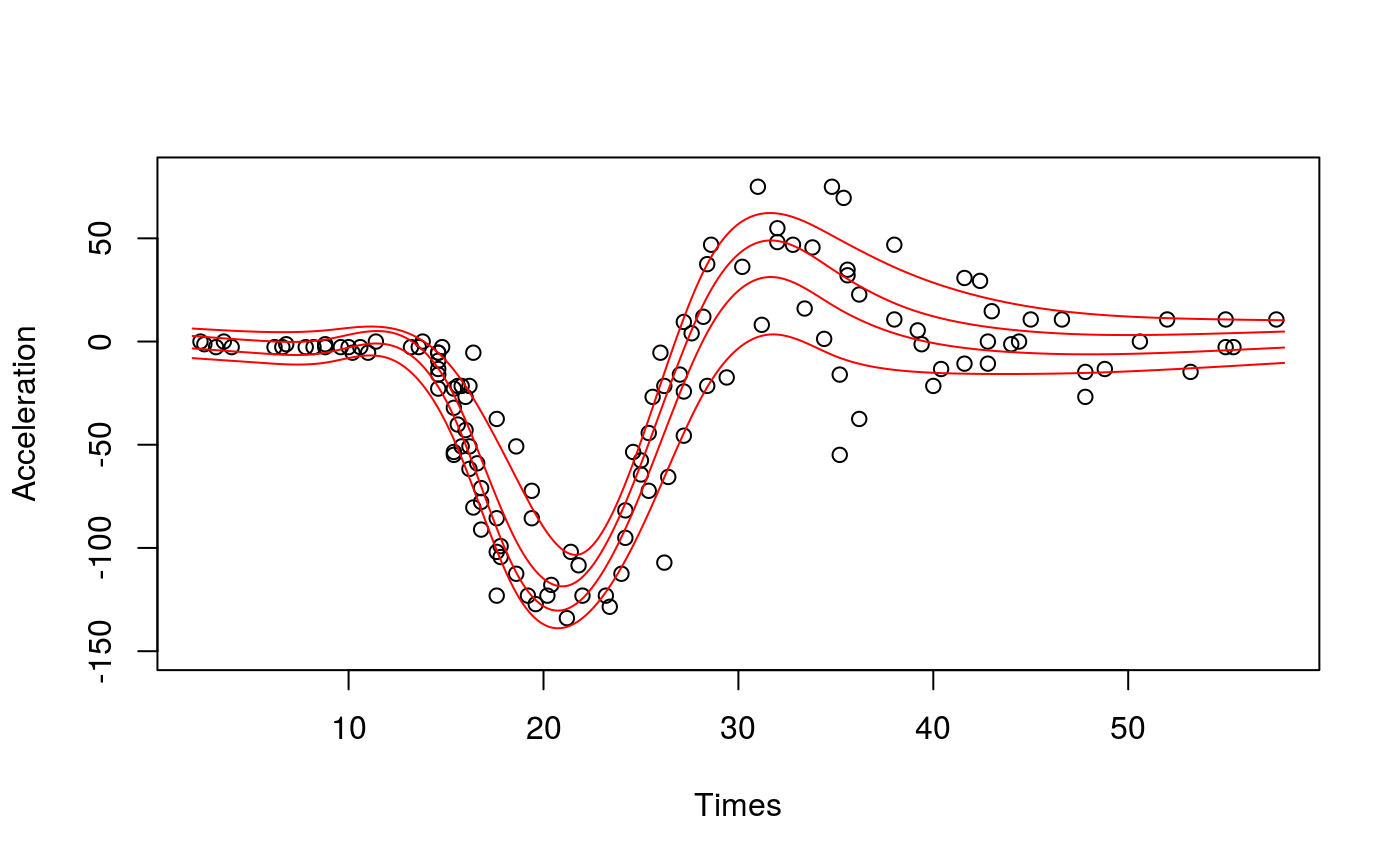
##### # Multivariate Gaussian example #### library(qgam) set.seed(2) dat <- gamSim(1, n=300, dist="normal", scale=2)#> Gu & Wahba 4 term additive model#> Estimating learning rate. Each dot corresponds to a loss evaluation. #> qu = 0.2...........done #> qu = 0.8..........done##### # Univariate "car" example #### library(qgam); library(MASS) # Fit for quantile 0.8 using the best sigma quSeq <- c(0.2, 0.4, 0.6, 0.8) set.seed(6436) fit <- mqgam(accel~s(times, k=20, bs="ad"), data = mcycle, qu = quSeq)#> Estimating learning rate. Each dot corresponds to a loss evaluation. #> qu = 0.4..........done #> qu = 0.6...........done #> qu = 0.2............done #> qu = 0.8..........done# Plot the fit xSeq <- data.frame(cbind("accel" = rep(0, 1e3), "times" = seq(2, 58, length.out = 1e3))) plot(mcycle$times, mcycle$accel, xlab = "Times", ylab = "Acceleration", ylim = c(-150, 80))