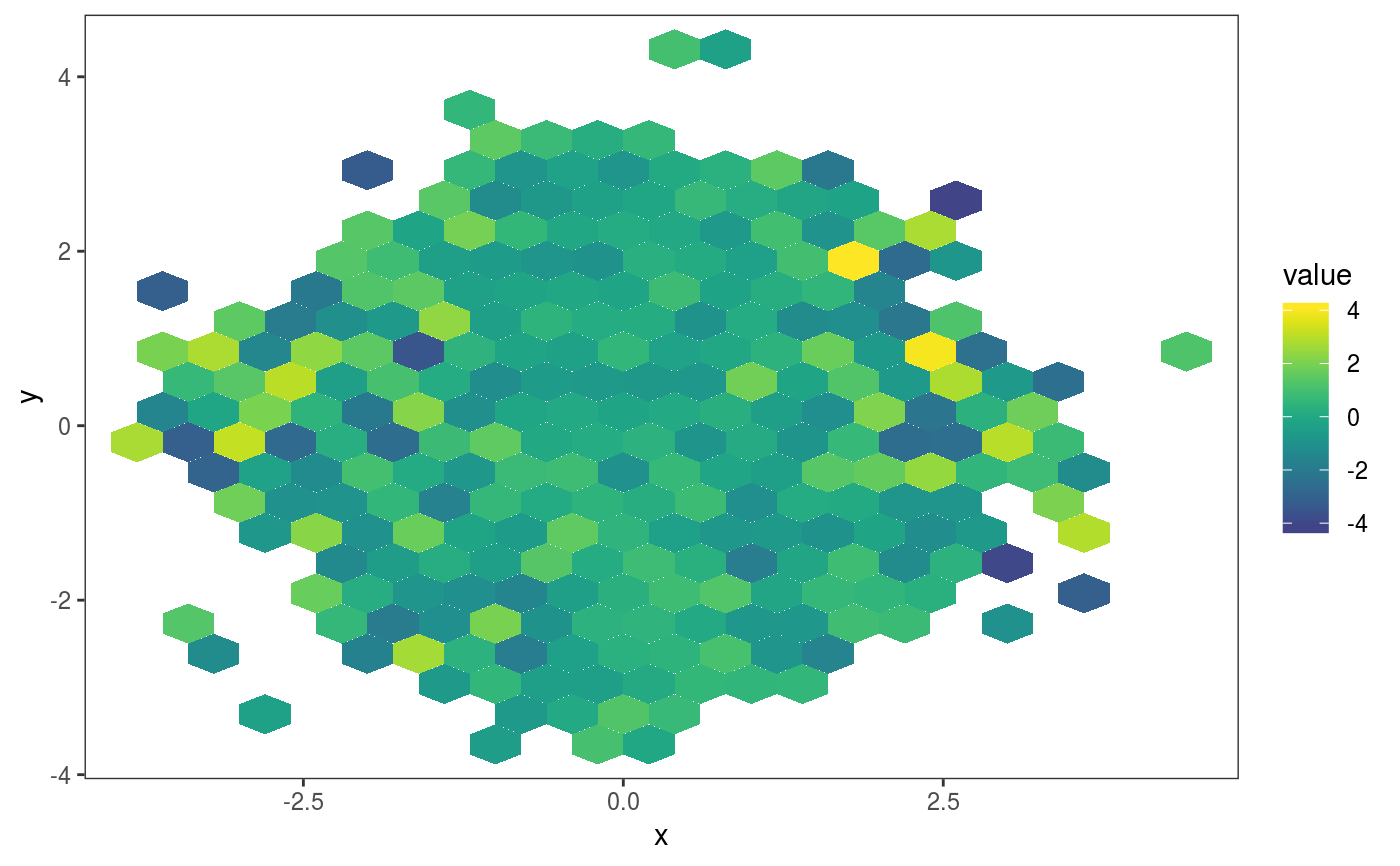
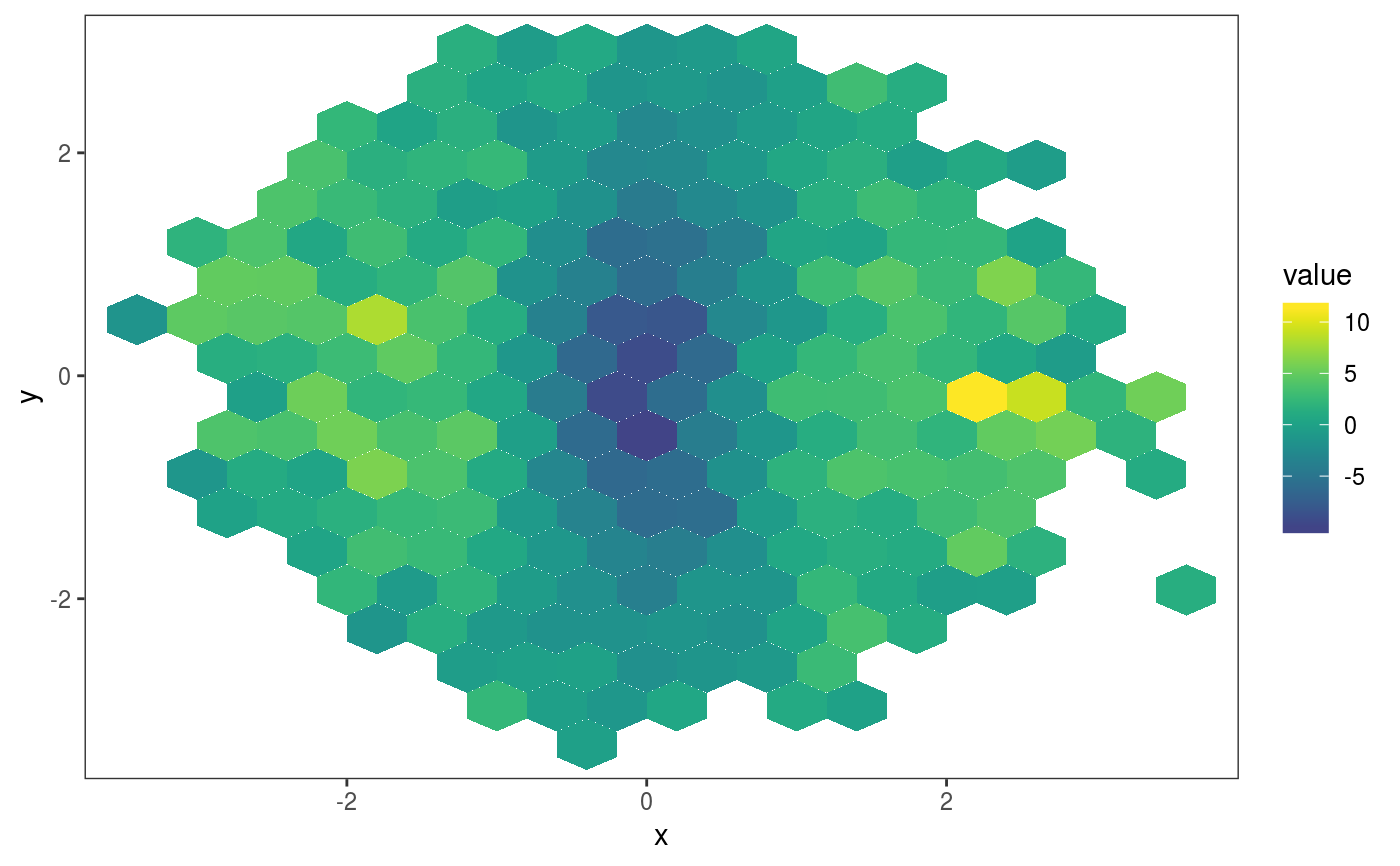
This layer bins the residuals, r, according to the value of the corresponding covariates, x1 and x2. Then the residuals in each bin are summarized using a scalar-valued statistic. Confidence intervals for the statistic corresponding to each bin can be obtained by simulating residuals from the fitted GAM model, which are then binned and summarized. Mainly useful in conjuction with check2D.
l_gridCheck2D(gridFun = mean, bw = c(NA, NA), stand = TRUE, binFun = NULL, ...)
Arguments
| gridFun | scalar-valued function used to summarize the residuals in each bin. |
|---|---|
| bw | numeric vector giving bin width in the vertical and horizontal directions. See the |
| stand | if left to |
| binFun | the |
| ... | graphical arguments to be passed to |
Value
An object of class gamLayer
Examples
library(mgcViz); set.seed(4124) n <- 1e4 x <- rnorm(n); y <- rnorm(n); # Residuals are heteroscedastic w.r.t. x ob <- (x)^2 + (y)^2 + (1*abs(x) + 1) * rnorm(n) b <- bam(ob ~ s(x,k=30) + s(y, k=30), discrete = TRUE) b <- getViz(b, nsim = 50) # Don't see much by looking at mean check2D(b, "x", "y") + l_gridCheck2D(gridFun = mean, bw = c(0.4, 0.4))# Variance pattern along x-axis clearer now check2D(b, "x", "y") + l_gridCheck2D(gridFun = sd, bw = c(0.4, 0.4))