This method can be used to simulate vectors of responses from the Gaussian posterior approximation of a gamObject.
postSim( o, nsim, newdata, trans = NULL, method = "auto", w = NULL, offset = NULL, savePar = FALSE, ... )
Arguments
| o | the output of a |
|---|---|
| nsim | the number of simulated vectors of responses. A positive integer. |
| newdata | Optional new data frame used to perform the simulations. To be passed to predict.gam. |
| trans | function used to transform or summarize each vector of simulated responses.
It must take a vector as argument, but it can output a vector or a scalar.
Potentially useful for saving storage (e.g. by transforming each simulated vector
to a scalar). If left to |
| method | the method used for the simulation of responses. See simulate.gam. |
| w | vector of prior weights of each response. See simulate.gam. |
| offset | numeric vector of offsets. For GAMs with multiple linear predictor (see eg gaulss) it
must be a list of vectors. If |
| savePar | if |
| ... | arguments to be passed to vcov.gam. |
Value
If savePar == FALSE the function will return a matrix where each row is a vector of
simulated responses or a transformed version of it. If savePar == TRUE it will return
a list where the $simY entry will contain the simulated responses and $simBeta
the simulated parameters.
Examples
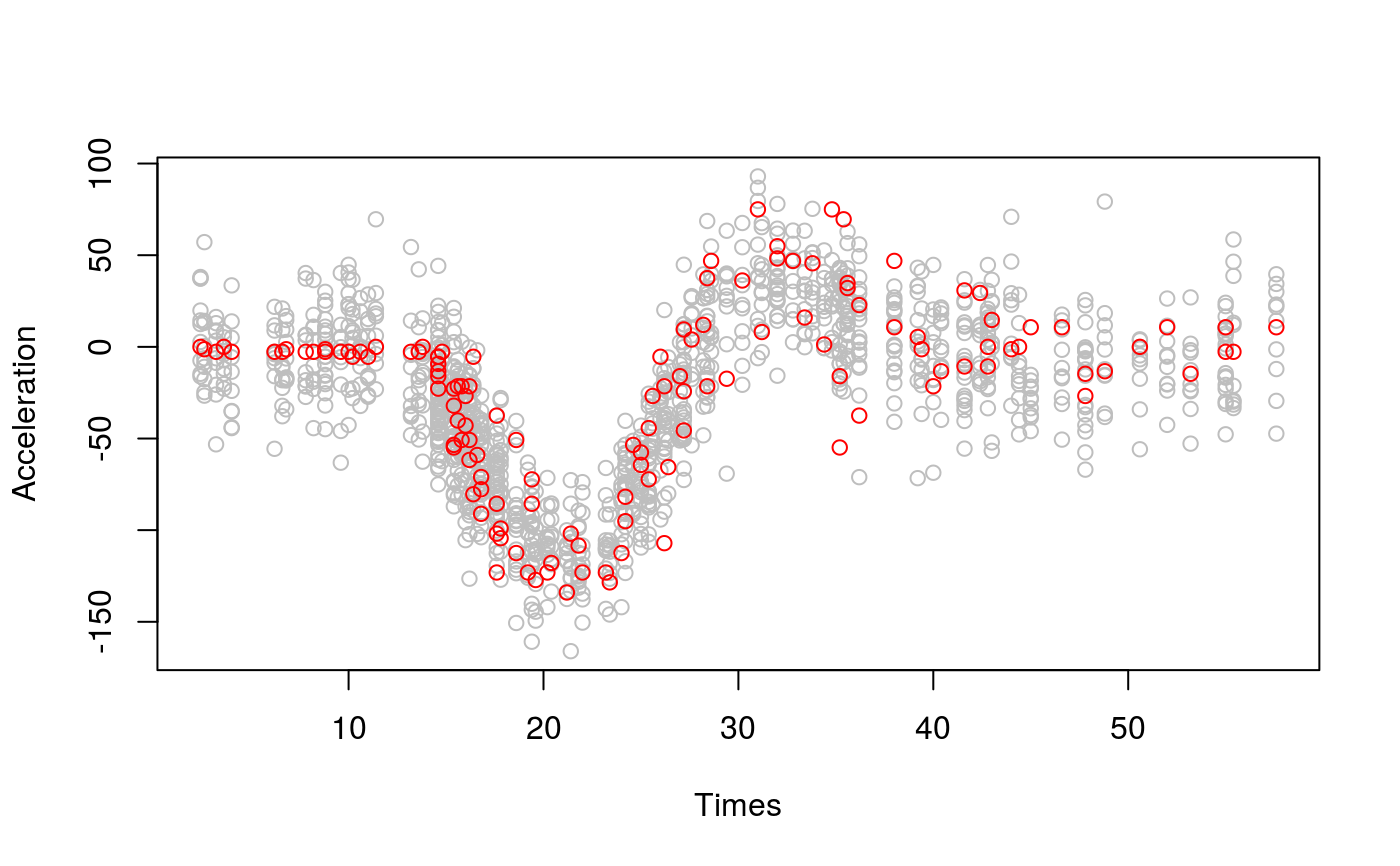
library(mgcViz) library(MASS) b <- gam(accel~s(times, k=20), data=mcycle) # Simulate list of 10 vectors of responses from posterior, taking into # account smoothing parameters uncertainty (see ?vcov.gam) n <- 10 sim <- postSim(o = b, nsim = n, unconditional = TRUE) # Posterior simulations in grey and data in red plot(rep(mcycle$times, n), as.vector(t(sim)), col = "grey", ylab = "Acceleration", xlab = "Times")# There is clear disagreement between simulations' and data's # conditional variance, which can be solved using flexible GAMLSS model: b <- gam(list(accel~s(times, k=20), ~s(times)), data=mcycle, family = gaulss) sim <- postSim(o = b, nsim = n) plot(rep(mcycle$times, n), as.vector(t(sim)), col = "grey", ylab = "Acceleration", xlab = "Times")